Prediction of gene regulatory networks
The regulation of transcription has been my primary research interest from the time when I was a graduate student. During my PhD course, I explored approaches for predicting gene expression from DNA sequence motifs, and higher-order properties of promoters, such as combinations of motifs, their positioning and orientation (see for example Vandenbon and Nakai, Nucl Acids Res, 2010).
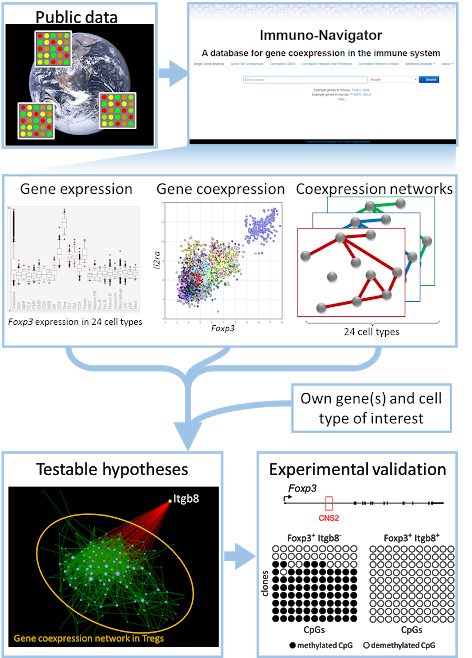
More recently, I constructed Immuno-Navigator, a gene co-expression database for cell types of the immune system (Vandenbon et al., PNAS, 2016). An important point of this study is that we thoroughly analyzed and reduced batch effects in our dataset, which improved its quality considerably. We successfully used the resulting co-expression data for predicting new regulators of importance specifically in Treg cells, in collaboration with the laboratory of Prof. Shimon Sakaguchi (Osaka University, link).
Analysis of changes in epigenetic markers after immune stimulation
Stimulation of immune cells using pathogen-associated molecular patterns induces not only changes in gene expression, but also in epigenetic markers at promoters and enhancer regions. These changes include acetylation and methylation of lysine residues in histone proteins. It remains unclear whether histone modifications play an active role in the regulation of transcription, or if they are mostly byproducts (see review by Henikoff and Shilatifard, Trends Genet., 2011). We have been studying the temporal order of changes in transcription and histone modifications in dendritic cells following lipopolysaccharide stimulation, using ChIP-seq data and bioinformatics (Vandenbon et al., Genome Biology, 2018). We are investigating causal relationships between transcription factors and the accumulation of histone modifications at LPS-induced promoters. This is a close collaboration with Dr. Yutaro Kumagai (IFReC, Osaka University; currently in AIST), and Profs. Yutaka Suzuki and Kenta Nakai (both in the University of Tokyo).
Interdisciplinary collaborations
I have participated in several collaborative research projects in immunology. For example, we have revealed interactions between transcription factors and epigenetic regulation, such as between Irf4 and Jmjd3 (Satoh et al., Nature Immunology, 2010), and between Jdp2 and inhibition of histone acetylation at the Atf3 promoter (Maruyama et al., Immunity, 2012). More recently, I contributed to the elucidation of the role of Akirin2 in recruiting the SWI/SNF complex to NF-κB target promoters (Tartey et al., EMBO J., 2014), and of our understanding of Treg-specific regulation of transcription, through the analysis of genome-wide Treg-specific DNA methylation patterns and the binding by Foxp3 (Morikawa et al., PNAS, 2014). On the level of post-transcriptional regulation of gene expression, through the integrative analysis of RIP-seq and gene expression data, we identified the target RNAs of the RNase Regnase-1 (Uehata et al., Cell, 2013; Mino et al., Cell, 2015). Downstream bioinformatics analysis of RNA sequence and structural motifs led to a better understanding of the RNA stem-loop structure which it recognizes.